🧬-Genomics and Reproducibility
Introduction
The study of genomics involves analyzing sequencing data to infer impactful biological findings, making replicability and transparency regarding how results are obtained paramount. Advancements in large-scale molecular biology methods have driven an unprecedented increase in data generation, pushing computational capacities to their limits and highlighting a growing need for robust and scalable data analyses. Consequently, Workflow Management Systems (WfMSs) are the recommended solution for dealing with high-throughput data analysis pipelines.
We explore core components necessary for implementing reproducible and scalable bioinformatics projects. Popular open-source WfMSs include Nextflow and Snakemake, which represent analyses as workflows where steps are wrapped into processes connected by data dependencies. These systems support core features critical for efficiency, such as automatic parallelization and portability across diverse infrastructures, including High-Performance Computing (HPC) clusters and cloud computing. Nextflow, currently experiencing the highest growth in usage among WfMSs, is the main driver behind their adoption.
To ensure analyses are independent of the execution machine, tool distribution must involve accurate tracking of dependencies and software environment versions. This challenge is addressed through containerization software such as Docker and Kubernetes, which allow the creation of encapsulated computer environments defined as images. This approach is the most comprehensive solution for dependencyi control, resulting in consistent execution across various operating systems (Linux, Windows, MacOS).
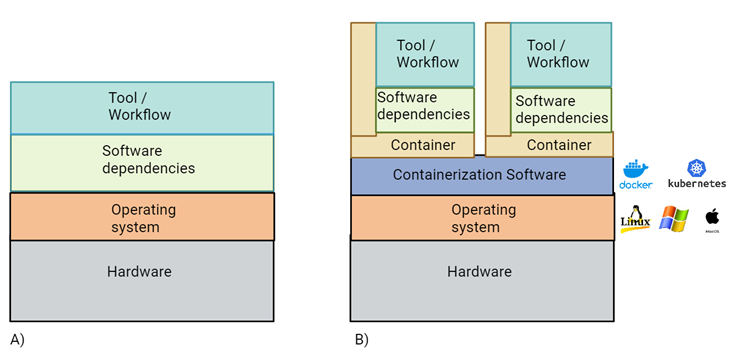
Figure 1 Representation of software containerization:
- A) Tools and related dependencies are installed system wide, possibly creating conflicts with future installation or software updates.
- B) Creation of containers using a containerization software allows to encapsulate software-specific dependencies, allowing different versions of the same libraries to be installed on the same operating system. Containerization allows creation of a reduced operating system with only the dependencies required to run the code. This results in consistent run on multiple infrastructures.
To maximize standardization, the nf-core framework provides a curated collection of community-developed pipelines based on agreed-upon best-practice standards. This framework leverages Nextflow’s Domain-Specific Language 2 (DSL2), a key technical advancement that allows complex workflows to be split into reusable modular components. These modules encapsulate the correct execution environment using container images, promoting improved code quality and long-term maintenance. The open-source model and use of platforms like Git and GitHub are central to enabling collaboration, version control, and increased software longevity.
Adopting these technologies enhances reproducibility and supports adherence to the FAIR principles (Findability, Accessibility, Interoperability, and Reusability). The benefits of using WfMSs and containerization include improving system-agnostic portability and enabling scalability from local environments to Cloud or HPC. WfMSs further produce accurate job logs, making analysis transparent.
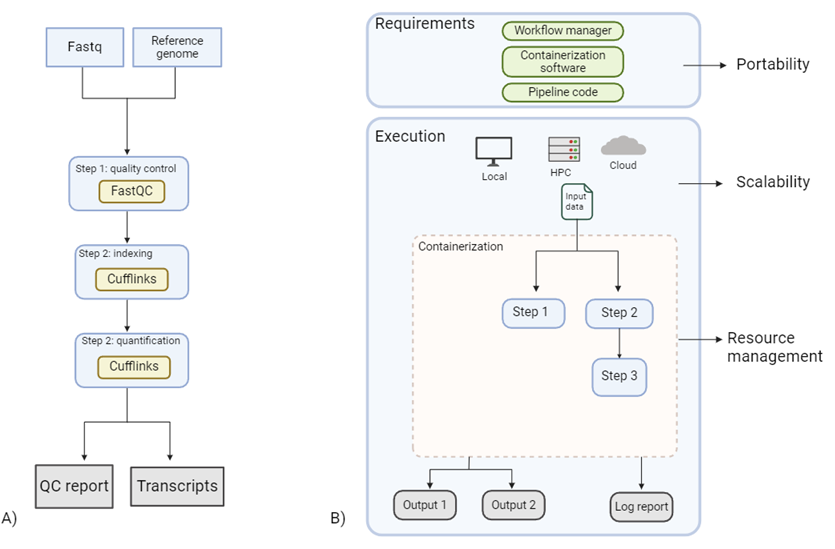
Figure 2 Improvements brought by workflow managers:
- A) A simple pipeline quantifying RNA transcript. Lack of workflow management and containerization makes the pipeline prone to breaking if correct dependencies are not installed or become unavailable.
- B) RNA quantification pipeline including a workflow manager, and containerization improving its system agnostic portability. Furthermore, scalability from local to Cloud or High-Performance Computing (HPC) is made possible via containerization. Finally, workflow managers allow parallelization of jobs automatically when possible, allowing efficient resource management. They further produced accurate job logs, making analysis transparent and reproducible.
In practice, the adoption of Nextflow, often in conjunction with nf-core standards, facilitates implementation of highly complex data analyses. The application within large distributed research groups, such as the EuroFAANG farmed animal genomics communities, demonstrates how a common best-practices analytic framework ensures the long-term comparability and standardization of results across time and multiple institutions. Nextflow’s language-agnostic property allows existing code (like Bash or Perl) to be ported with limited refactoring, enabling a progressive standard transition and the deployment of execution at scale in a portable and replicable manner across heterogeneous execution platforms. Courses like the Reprohackathon also highlight that implementing reproducible analyses is a complex task, but training in these concepts and tools significantly improves researchers’ abilities in this domain.